Bioanalytical Research
Bioanalytical Research Group

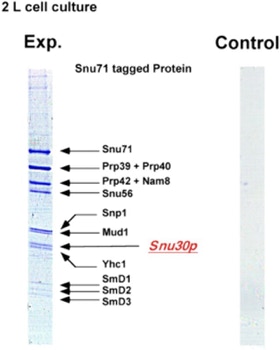
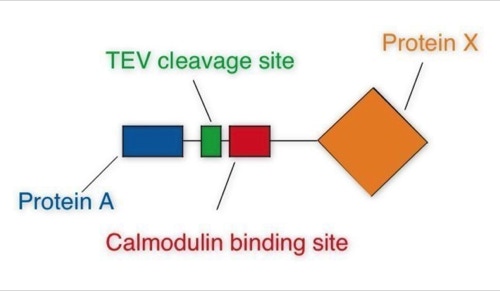
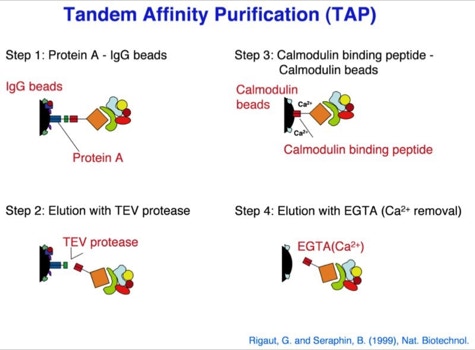
One of the early important decisions in 1999 was to dedicate all available resources to the support of the “Tandem Affinity Purification” (TAP) method [1]. The TAP method had been developed in Bertrand Seraphin’s group. It was a tag based, gentle, specific and in-vivo protein complex purification method established in yeast cells (slide 2, 3). This technology could evolve together with mass spectrometry based protein identification into a key component of an organism wide functional proteomics projects - a systematic effort to place proteins into their functional cellular context (slide 4, 6) [2].
Later we demonstrated how the TAP method could be used with cells from higher eukaryotes, the iTAP project (slide 7, 8, 9) [3].
The Tandem Affinity Purification Method:
The TAP method was so important because it allowed to purify non-covalent protein complexes in a systematic fashion with high specificity without requiring individual antibodies for every complex (slide 1, 2, 3). Protein complexes are assemblies of proteins who cooperate to fulfil a specific function within the cell. At a time when the human genome was going to be published this technique would allow to place many of the newly discovered proteins into their functional context. This activity was supported by the German Bundesministerium für Bildung und Forschung (BMBF) (BioFuture, Grant 0311862 / 5).
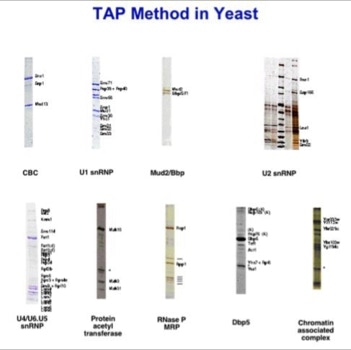
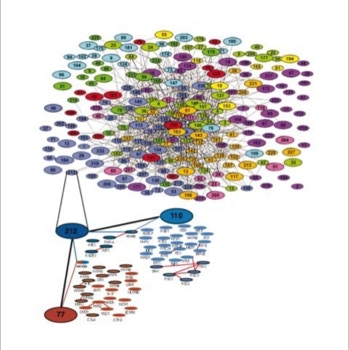
The success of this project at the EMBL motivated a proteomics spin-off company, CellZome, to use it as their core activity (slide 5, 6). They could generate the first proteome wide protein complex network [4].
Using the TAP Method in Cells from Higher Eukaryotes - the iTAP Project:
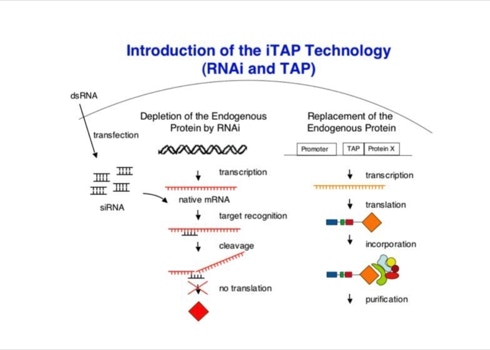
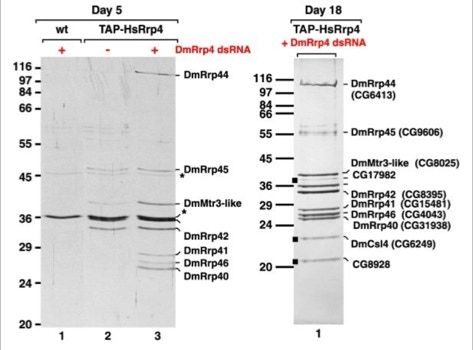
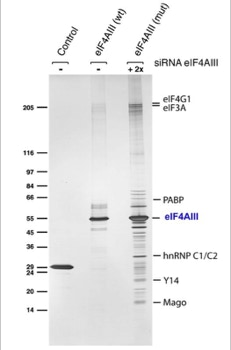
The first attempts to use TAP in Drosophila cells were a failure. Expressing a tagged protein and extracting it was often not enough to allow a specific and efficient purification of its interacting partners. This caused us to modify the experiments. Expression of the TAP-tagged protein was accompanied by suppression of its native form using interference RNA (RNAi). RNAi is a method that allows to hydrolize the native messenger RNA of a protein so that it can not be synthesized (slide 7). With the native protein missing the tagged one should be built into the biological pathways. Upon purification its native partners should be extractable. The biochemical experiments were done in the group of Elisa Izaurralde. For the protein complexes purified the project was a complete success (slide 8, 9) [3]. The iTAP method was incorporated into a patent.
iTAP Methods - Experimental Information:
- Plasmids
- Drosophila cell culture
- Generating stable cell lines
- RNAi protocol
- IgG crosslinking
- Buffers
- Tandem Affinity Purification
- Protein separation and staining
References*
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17, 103
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. (2001). The tandem affinity purification (tap) method: a general procedure of protein complex purification. Methods 24, 218-29
- Forler, D., Kocher, T., Rode, M., Gentzel, M., Izaurralde, E., and Wilm, M. (2003). An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotechnol 21, 89-92.
- Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., Remor, M., Hofert, C., Schelder, M., Brajenovic, M., Ruffner, H., Merino, A., Klein, K., Hudak, M., Dickson, D., Rudi, T., Gnau, V., Bauch, A., Bastuck, S., Huhse, B., Leutwein, C., Heurtier, M. A., Copley, R. R., Edelmann, A., Querfurth, E., Rybin, V., Drewes, G., Raida, M., Bouwmeester, T., Bork, P., Seraphin, B., Kuster, B., Neubauer, G., and Superti-Furga, G. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141-7.
* It is not intended that the reference list reflects the entire research in the field.