Bioanalytical Research

Sequence & Identify Proteins
Bioanalytical Research Group
In 1992 a mass spectrometry group was established under the leadership of Matthias Mann. In the following years we explored how to sequence peptides and identify proteins using mass spectrometers in the most sensitive way. There were three important components to this
• the nano-electrospray ion source (figure 1 - 4)
• the sequence tag based protein identification algorithm (slide 5)
• protocols for gel staining, in-gel digestion and sample preparation for the analysis with mass spectrometry
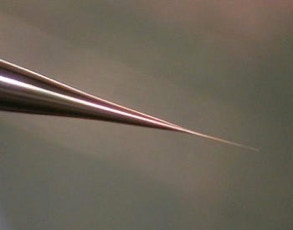

Figure 1: Nano-Electrospray Emitter
Figure 2: Nano-Electrospray Ion Source
The nano-electrospray ion source:
A theoretical and experimental study of the electrospray process done in 1988 / 1989 at the University of Münster in the department of experimental physics in the group of Prof. Benninghoven (by Matthias Wilm) let to the construction principle of the nano electrospray ion source [1].
The result was that an electrospray emitter produces exclusively droplets with a diameter of 200 nm or less when operated at a flow rate of 20 nL/min or less. The spray should be generated purely electrostatically emitting the droplets from a stable Taylor Cone. The small size is so important because every droplet contains on the average only one analyte molecule per droplet if the analyte concentration is in the order of 1 pmol/µl. The nano electrospray ion source could therefore represent an electrospray ion source with a 100% ionization efficiency [1, 2]. At its time these findings proposed a mechanism for the electrospray ionization effect itself. The molecules would be passively released into the vacuum system after evaporation of the solvent. They allowed the prediction that the electrospray ionization mechanism is a very soft mechanism and that it should be possible to ionize molecules or molecular assemblies of virtually unlimited mass. The experimental results at that time were already produced by an electrospray emitter made from pulled glass capillaries.
The hoped for characteristics of the later called nano-electrospray ion source could be realized in the early 90's at the EMBL in the group of Matthias Mann. The pulled glass capillaries are the key element of the ion source. They were produced with a micro-capillary puller. Their surface is covered with gold or any other metal to make them conductive. The needles are mounted in gas tight needle holder to allow the application of a small air pressure for supporting the constant flow of liquid through the narrow tip if required. When coming from the puller the orifice of the needles can be in the nanometers range. For stable operation they have to be opened to give an orifice of 1 - 2 µm by moving them gently against a surface under microscopic control. Since nano-electrospray needles when operated on flow rates of 10 - 20 nL / min produce exclusively very small droplets they can be directly interfaced with the vacuum system of a mass spectrometer. The short distance to the orifice of the mass spectrometer, its high ionization efficiency and the low flow rate makes the nano-electrospray source a very useful emitter for the analysis of individual samples for direct mass measurements or tandem mass spectrometric investigations [2].
The nano electrospray source had been made commercially available for a variety of instruments by Proxeon.


Figure 3: Nano-Electrospray Emitter
Figure 4: Puller Equipment to Generate Nano-Electrospray Emitters
Sequence tag based protein identification:
The sequence tag based protein identification algorithm is a manual way to identify protein based on the tandem mass spectrum of one or several of its peptides [3]. The algorithm was invented by Matthias Mann at a time when low resolved triple quadrupole fragment spectra were the only source of sensitive fragment spectra of peptides. A sequence tag is a short stretch of sequence read out from a fragment spectrum of a peptide.
In the example of slide 5 which shows the fragment spectrum of the peptide TTPAVLDSDGSYFLYSK the sequence tag is (1079.61)SD(IL)(1394.80). The amino acids I and L can not be distinguished in a low energy fragment spectrum. To lead to the correct identification of the underlying peptide the sequence tag has to connect a continuous series of b or (exclusive or) y ions. If a peptide had been found via a database search the complete peptide sequence is compared with the complete fragment spectrum to find other sequence specific ions for confirming the identification. The sequence tag based identification algorithm is a very specific identification method.
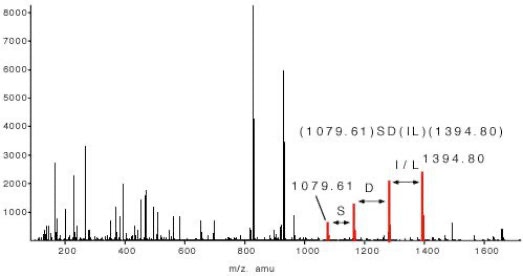
Figure 5: Sequence Tag
Sample preparation protocols:
Several protocols relevant for the analysis of proteins by mass spectrometry had been used in our group. The silver staining and the in-gel digestion protocol had been developed by Andrej and Anna Shevchenko [4].
• Mass spectrometry compatible silver staining of gels: silver staining protocol
• In-gel digestion of proteins to be analyzed by mass spectrometry: in-gel digestion protocol
• Two methods for the sample preparation for MALDI: dried droplet, fast evaporation protocol
• A selection of MALDI matrices: MALDI matrices
By introducing 18O labelling it became possible to sequence long amino acid stretches from peptide fragment spectra de novo [5].
References*
- Wilm, M. and M. Mann, Electrospray and Taylor-Cone Theory, Dole's Beam of Macromolecules at last ? International Journal of Mass Spectrometry and Ion Processes, 1994. 136: p. 167 - 180.
- Wilm, M., and Mann, M. (1996). Analytical Properties of the Nanoelectrospray Ion Source. Analytical Chemistry 68, 1 - 8.
- Mann, M., and Wilm, M. (1994). Error-Tolerant Identification of Peptides in Sequence Databases by Peptide Sequence Tags. Analytical Chemistry 66, 4390 - 4399.
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass Spectrometric Sequencing of Proteins from Silver - Stained Polyacrylamide Gels. Analytical Chemistry 68, 850 - 858.
- Shevchenko, A., Chernushevich, I., Ens, W., Standing, K. G., Thomson, B., Wilm, M., and Mann, M. (1997). Rapid 'de Novo' Peptide Sequencing by a Combination of Nanoelectrospray, Isotopic Labeling and a Quadrupole/Time-of flight Mass Spectrometer. Rapid Communications in Mass Spectrometry 11, 1015 - 1024.
* It is not intended that the reference list reflects the entire research in the field.